Information ecology and the evolution of complexity

‘Open-ended increase in complexity’ is a thing that we often want to find or make in ALife. The idea that we could come up with some simulation that, if we just threw more time and computer power at it, would constantly reveal new things to us is a bit of a holy grail. It also seems to be something that life on Earth has at first glance managed to do (although there have been bursting periods and stagnant periods). But in general, if you visited Earth 2 billion years ago, there would be a large number of biological innovations that had yet to come about despite the fact that there had been life on Earth for quite some time up to that point. 2 billion years is a very long time to go (in terms of generations of cells) without realizing that you’ve done everything there is to do.
It seems to be pretty difficult to get, though. Evolution alone isn’t enough – you can find all sorts of simulations with evolution where it happily goes along, discovering new things, until it finds that it has discovered the best thing ever – the best possible replicator, the most competitive genome, the optimal solution to the problems posed by the simulation. So once it finds that, the system just sits there doing that thing for the rest of time. Alternatively, there are simulations where a fluctuating environment or competitive cycle causes the organisms to constantly change what they’re doing. But when you look carefully, you find that the system is repeating, or just doing things randomly (so that it is in some sense statistically stationary). Or, a more subtle thing, it might even go off and make something arbitrarily elaborate and complex but which remains qualitatively identical – it makes all sorts of elaborate trees, but it never makes something that isn’t a tree.
When someone first starts doing ALife and is first faced with this conundrum, there’s a sequence of ideas that almost everyone goes through. ‘Maybe we just need to run it longer?’, ‘Maybe the system just needs to be bigger?’, ‘What if you add a fluctuating environment?’, etc. These are all pretty easy to try out, and so one quickly explores these possibilities and builds an intuition about what each of these things is doing (hopefully). But there’s a much harder one to answer which keeps coming up: ‘What if, to get a lot of complexity in life, we need a lot of complexity in the laws of physics/the world?’.
This is a particularly tricky thing, because its one of those hypotheses that if it’s really the answer to the question, it shuts down the purpose of the whole endeavor. If we can only get out what we put in, why bother at all? It also looks counter-intuitive given what we see in the real world – chemistry has a lot of complexity and structure, but it’s a natural result of quantum mechanics which has much less complexity ‘baked in’. There are many more chemical compounds than chemical elements, and this sort of combinatoric character feels like it should get you something. We also see things like programming languages, where a program can be much more complex than the compiler used to generate it.
Even if the idea feels unsatisfying, it would be good to give it a fair shake and see whether or not there’s anything to it. Maybe rather than just getting out what we put in, there’s a sort of leverage where we get out something proportional to but greater than what we put in, which would give us an idea of how to design the kinds of feedbacks we would need to chain into real open-endedness. Or there could be some other subtlety revealed in careful examination. And if we found that actually complexity in the environment doesn’t always get you something, it would suggest that we can maybe put this particular direction to rest and look elsewhere for the time being.
Its not enough to just pick a favorite system, throw it into a complex environment, and see what happens – if you got a null result, you wouldn’t be able to say that it wasn’t just a bad choice, and that it doesn’t say anything in general. Instead what I’m going to try to do here is to come up with a system where I can guarantee that an increase in environmental complexity results in an increase in the complexity of the resulting evolutionary system – the hope being that then if it fails, it suggests at the least that there should be some reason why it wasn’t possible to build that might point at a deeper understanding. Towards this end, perhaps a place to start is to look at deep learning, where we know that large amounts of rich data do make an observable difference in the way neural networks learn. In such cases, the neural networks discover statistical and structural regularities in the data that begin to let them actually generalize to widely different problems – a network pre-trained to recognize what sport is being played in a video is actually faster at learning to recognize objects than starting from scratch.
If I want to make an evolutionary analogue of this, I think the important thing is to recognize that once a neural network correctly classifies a given piece of data, it can be shown that data again without having its weights change. So it naturally insulates the parts of it that have already learned things from the expanding fronts where there are still new things to learn. An ecological version of this is the concept of niches – if I have some organisms adapted to one niche, they don’t compete with what’s going on in a different niche.
So what if we treat each part of a set of real data as corresponding to a separate niche? In that case, we’d at least find that as the data becomes more complex, the number of niches increases, and so the complexity of the resultant ecology would increase. That’s very schematic, but its enough to consider how to actually design such a system.
If I have different independently provisioned and limited food sources, such that an organism must decide which food source to eat from, then that immediately gives me a number of independent niches equal to the number of food sources. So given some data set, I can treat each measured feature as a food source, and then allow the organisms to try to predict that feature using some other features. The result would be that each feature is like a separate evolutionary simulation in total isolation from the others, where it’s just trying to find the best way to predict that one target. However, for it to really qualify as complexity and not just ‘large amounts of stuff’, I want these things to all interact with each other somehow.
The way I’ll try to do that is to make it so that any organism which reads a certain feature to use in its predictions must pay some resources into that bin. So the population capacity of a given feature is based on how useful that feature ends up being for predicting other things about the data down the line. Features which are highly predictive will end up being worth more if you can in turn predict them. The result would conceivably some kind of explanatory network, where for any given feature you can determine different ways to infer that feature given other sparse measurements. For example, if the network discovered that features A and B predict feature C, and D and E predict B, then you can figure out that if you knew D and A and wanted to predict C, you can measure either B or E and propagate the result through the network.
In terms of the specific implementation, I associated every feature of the data set with a small food supply. Organisms can pull from that food supply based on how their predictions of that feature compare with their competitors, so each feature is its own ecological niche. That means that at least for the simple one organism = one prediction case, we should expect to see a number of stable species more or less equal to the number of features in the data set (if organisms could generate new features, we’d have a model with niche creation, but in this simple version they cannot). When organisms have eaten a certain amount of food, they replicate. This is balanced against all organisms having a constant death rate. For the predictor, each organism has a small neural network which is trained during its lifetime. However, the inputs, outputs, and hidden layer of the neural network are all specified genetically. Mutation occurs during replication, and alters the parameters of the network.
I haven’t gone into a full description of the model or the algorithm, and to be honest there are a lot of knobs to twist and things that I had to tweak to get something without obvious ‘cheating’ behavior (for example, if someone figures out how to use a feature to predict that same feature). The full layout would be a bit much for this blog post, but I’ve made some source-code available (it uses Python, with Lasagne/Theano for the neural nets) with example data from the survivors of the Titanic.
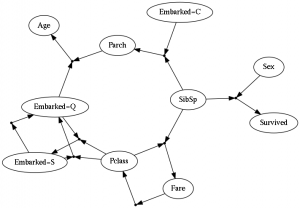
This setup does appear to work, at least in the sense that the evolutionary dynamics do increase the average predictive power of the collective population over time. You get some interesting networks that might suggest all sorts of potential relationships between features of the data, so this could also be useful for feature engineering. It’s unclear whether this is really ‘complex’ though, or if its just a lot of different independent things. The lack of any form of niche creation seems to severely limit how interesting the results can get. Presumably we’d want organisms to be able to use a hidden neuron to ‘summarize’ some collection of features about the data, and then perhaps have some other organism come along and make use of the hidden neuron itself rather than the base features in order to save on costs – e.g. creating something like a food web or series of informational trophic levels. These new features wouldn’t have an influx of food associated with them, but could gain associated food if the end up being useful to other organisms. So the result would be that the number of niches would start at being the number of features, but it could become larger if it is somehow useful to do so in order to more efficiently explain the data.
This might start to get at the original question: does something like this just mirror the complexity of the data set, or is there some way to leverage that initial complexity to generate something with greater complexity (as well as greater explanatory efficiency) than before. Certainly such a thing would not go open-ended, but it might give a good way to really provide environmental complexity that gets used by the resulting ecology, rather than simply being integrated over or ignored.